-Search query
-Search result
Showing 1 - 50 of 96 items for (author: locke & j)
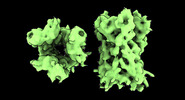
EMDB-17704: 
Subtomogram average of Vaccinia A10 trimer with open center from in vitro cores
Method: subtomogram averaging / : Turonova B, Liu J
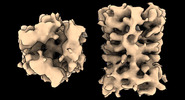
EMDB-17708: 
Subtomogram average of Vaccinia A10 trimer with tight center from in vitro cores
Method: subtomogram averaging / : Turonova B, Liu J

EMDB-17753: 
Subtomogram average of Vaccinia A10 trimer from in situ cores
Method: subtomogram averaging / : Turonova B, Liu J

EMDB-27781: 
Cryo-EM structure of 227 Fab in complex with (NPNA)8 peptide
Method: single particle / : Martin GM, Ward AB

EMDB-27784: 
Cryo-EM structure of 239 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

EMDB-27785: 
Cryo-EM structure of 311 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

EMDB-27786: 
Cryo-EM structure of 334 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

EMDB-27787: 
Cryo-EM structure of 337 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

EMDB-27788: 
Cryo-EM structure of 356 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

EMDB-27789: 
Cryo-EM structure of 364 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

PDB-8dyt: 
Cryo-EM structure of 227 Fab in complex with (NPNA)8 peptide
Method: single particle / : Martin GM, Ward AB

PDB-8dyw: 
Cryo-EM structure of 239 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

PDB-8dyx: 
Cryo-EM structure of 311 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

PDB-8dyy: 
Cryo-EM structure of 334 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

PDB-8dz3: 
Cryo-EM structure of 337 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

PDB-8dz4: 
Cryo-EM structure of 356 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

PDB-8dz5: 
Cryo-EM structure of 364 Fab in complex with recombinant shortened Plasmodium falciparum circumsporozoite protein (rsCSP)
Method: single particle / : Martin GM, Ward AB

EMDB-14860: 
Cryo-EM structure of the MVV CSC intasome at 4.5A resolution
Method: single particle / : Ballandras-Colas A, Maskell D, Pye VE, Locke J, Swuec S, Kotecha A, Costa A, Cherepanov P

PDB-7zpp: 
Cryo-EM structure of the MVV CSC intasome at 4.5A resolution
Method: single particle / : Ballandras-Colas A, Maskell D, Pye VE, Locke J, Swuec S, Kotecha A, Costa A, Cherepanov P

EMDB-13176: 
3.0 A resolution structure of a DNA-loaded MCM double hexamer
Method: single particle / : Greiwe JF, Locke J, Nans A, Costa A

EMDB-13211: 
Structure of a DNA-loaded MCM double hexamer engaged with the Dbf4-dependent kinase
Method: single particle / : Greiwe JF, Locke J, Nans A, Costa A

PDB-7p30: 
3.0 A resolution structure of a DNA-loaded MCM double hexamer
Method: single particle / : Greiwe JF, Miller TCR, Martino F, Costa A

PDB-7p5z: 
Structure of a DNA-loaded MCM double hexamer engaged with the Dbf4-dependent kinase
Method: single particle / : Greiwe JF, Miller TCR, Martino F, Costa A

EMDB-11222: 
Structure of SARS-CoV-2 spike glycoprotein (S) trimer determined by sub-tomogram averaging
Method: subtomogram averaging / : Turonova B, Sikora M, Schurmann C, Hagen W, Welsch S, Blanc FEC, von Bulow S, Gecht M, Bagola K, Horner C, van Zandbergen G, Landry J, de Azevedo NTD, Mosalaganti S, Schwarz A, Covino R, Muhlebach M, Hummer G, Locker JK, Beck M

EMDB-11223: 
Structure of SARS-CoV-2 spike glycoprotein (S) monomer in a closed conformation determined by sub-tomogram averaging
Method: subtomogram averaging / : Turonova B, Sikora M, Schurmann C, Hagen WJH, Welsch S, Blanc FEC, von Bulow S, Gecht M, Bagola K, Horner C, van Zandbergen G, Landry J, de Azevedo NTD, Mosalaganti S, Schwarz A, Covino R, Muhlebach M, Hummer G, Locker JK, Beck M

EMDB-11338: 
Kinesin binding protein (KBP)
Method: single particle / : Atherton J, Hummel JJA, Olieric N, Locke J, Pena A, Rosenfeld SS, Steinmetz MO, Hoogenraad CC, Moores CA

EMDB-11339: 
Kinesin binding protein complexed with Kif15 motor domain
Method: single particle / : Atherton J, Hummel JJA, Olieric N, Locke J, Pena A, Rosenfeld SS, Steinmetz MO, Hoogenraad CC, Moores CA

EMDB-11340: 
Microtubule complexed with Kif15 motor domain. Symmetrised asymmetric unit
Method: single particle / : Atherton J, Hummel JJA, Olieric N, Locke J, Pena A, Rosenfeld SS, Steinmetz MO, Hoogenraad CC, Moores CA

PDB-6zpg: 
Kinesin binding protein (KBP)
Method: single particle / : Atherton J, Hummel JJA, Olieric N, Locke J, Pena A, Rosenfeld SS, Steinmetz MO, Hoogenraad CC, Moores CA

PDB-6zph: 
Kinesin binding protein complexed with Kif15 motor domain
Method: single particle / : Atherton J, Hummel JJA, Olieric N, Locke J, Pena A, Rosenfeld SS, Steinmetz MO, Hoogenraad CC, Moores CA

PDB-6zpi: 
Microtubule complexed with Kif15 motor domain. Symmetrised asymmetric unit
Method: single particle / : Atherton J, Hummel JJA, Olieric N, Locke J, Pena A, Rosenfeld SS, Steinmetz MO, Hoogenraad CC, Moores CA

EMDB-11347: 
Structure of SARS-CoV-2 spike glycoprotein (S) trimer with one receptor binding domain (RBD) in open-state determined by subtomogram averaging
Method: subtomogram averaging / : Turonova B, Sikora M, Schurmann C, Hagen W, Welsch S, Blanc FEC, von Bulow S, Gecht M, Bagola K, Horner C, van Zandbergen G, Laundry J, de Azevedo NTD, Mosalaganti S, Schwarz A, Covino R, Muhlebach M, Hummer G, Locker JK, Beck M
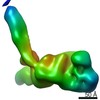
EMDB-10870: 
Cohesin complex with loader gripping DNA
Method: single particle / : Higashi TL, Eickhoff P, Sousa JS, Costa A, Uhlmann F

EMDB-10930: 
Cohesin complex with loader gripping DNA
Method: single particle / : Higashi TL, Eickhoff P, Sousa JS, Costa A, Uhlmann F

PDB-6yuf: 
Cohesin complex with loader gripping DNA
Method: single particle / : Higashi TL, Eickhoff P, Sousa JS, Costa A, Uhlmann F

EMDB-10744: 
Microtubule Nucleation by Single Human gamma-TuRC in a Partly Open Asymmetric Conformation
Method: single particle / : Locke J, Costa A

EMDB-10421: 
Human kinesin-5 motor domain in the GSK-1 state bound to microtubules
Method: helical / : Pena A, Sweeney A, Cook AD, Moores CA, Topf M

PDB-6ta3: 
Human kinesin-5 motor domain in the GSK-1 state bound to microtubules (Conformation 1)
Method: helical / : Pena A, Sweeney A, Cook AD, Moores CA, Topf M

EMDB-20444: 
Fab667 in complex with recombinant, shortened circumsporozoite protein
Method: single particle / : Torres JL, Ward AB

EMDB-20445: 
Fab668 in complex with recombinant, shortened circumsporozoite protein
Method: single particle / : Torres JL, Ward AB

EMDB-10422: 
Human kinesin-5 motor domain in the AMPPNP state bound to microtubules
Method: helical / : Pena AP, Sweeney A, Cook AD, Moores CA, Topf M

PDB-6ta4: 
Human kinesin-5 motor domain in the AMPPNP state bound to microtubules
Method: helical / : Pena AP, Sweeney A, Cook AD, Moores CA, Topf M

PDB-6tiw: 
Human kinesin-5 motor domain in the GSK state bound to microtubules (Conformation 2)
Method: helical / : Pena A, Sweeney A, Cook AD, Moores CA, Topf M

EMDB-20772: 
Fab397 in complex with NPNA8 peptide (Class 1)
Method: single particle / : Ward AB, Torres JL

EMDB-20773: 
Fab397 in complex with NPNA8 peptide (Class 2)
Method: single particle / : Ward AB, Torres JL

EMDB-20774: 
Fab397 in complex with NPNA8 peptide (Class 3)
Method: single particle / : Ward AB, Torres JL
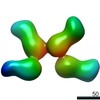
EMDB-20775: 
Fab397 in complex with rsCSP (Class 1)
Method: single particle / : Ward AB, Torres JL

EMDB-20776: 
Fab397 in complex with rsCSP (Class 2)
Method: single particle / : Ward AB, Torres JL
EMDB-20777: 
Fab397 in complex with rsCSP (Class 3)
Method: single particle / : Ward AB, Torres JL
EMDB-20778: 
Fab397 in complex with rsCSP (Class 4)
Method: single particle / : Ward AB, Torres JL
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
